University of Maryland (UMD) and U.S. Food and Drug Administration (FDA) researchers are working together to develop a new X-ray based technique that has the potential to advance understanding of Alzheimer’s disease in a preclinical stage. The group’s immediate aim is to develop a noninvasive way to quickly measure the amount of amyloid plaque in the brain, which has been found to correspond with Alzheimer’s disease progression. The researchers’ work was recently published in Scientific Reports.
In a healthy human brain, billions of specialized cells called neurons send information to one another via electrical and chemical signals in order to coordinate virtually all the body’s functions. Alzheimer’s disease disrupts these lines of communication, leading to a loss of function and cell death. These disruptions typically occur first in parts of the brain linked to memory, but over time, they affect areas that control language, reasoning, communication, mobility and other vital functions for living.
Scientists have long linked the accumulation of a protein fragment—beta-amyloid—in the brain to Alzheimer’s disease. While there are still many questions surrounding what exactly causes beta-amyloid to form plaques in the brain, the ability to effectively measure this buildup could shed new light on disease progression and therapeutic approaches in patients.
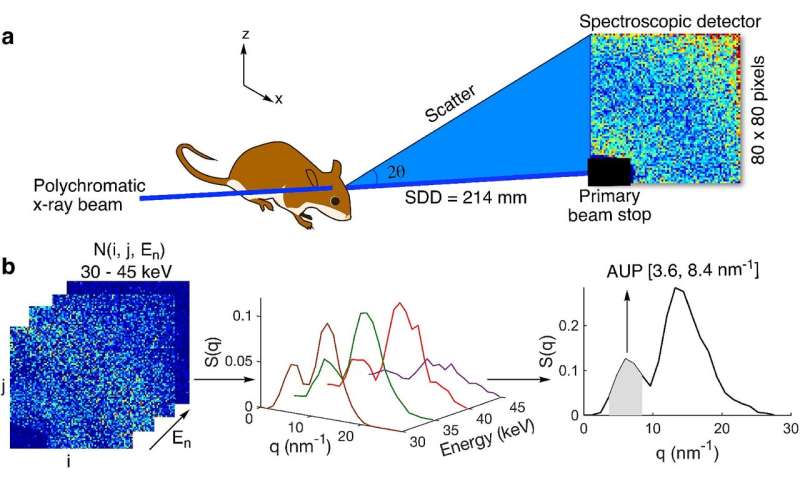
“In this work, we used spectral X-ray techniques to capture the coherent X-ray scattering characteristic of beta-amyloid aggregates deep in the brain without the need for injecting imaging agents,”said Fischell Department of Bioengineering (BIOE) adjunct professor Aldo Badano, corresponding author on the Scientific Reports paper. In addition to his BIOE appointment, Badano is a Deputy Director for the Division of Imaging, Diagnostics and Software Reliability with the FDA’s Center for Devices and Radiological Health (CDRH) in the Office of Science and Engineering Laboratories (OSEL).
Current methods to measure this amyloid burden face significant shortcomings. Studies of amyloid burden in Alzheimer’s disease are either limited to post-mortem analysis or they require use of resource-intensive PET or MRI scans that require injection of radiotracers or contrast agents. In addition, current methods for studying amyloid burden in vivo lack the ability to deliver images with high enough specificity and resolution to enable a thorough understanding of disease progression.
Recognizing this, Badano joined forces with BIOE alum Eshan Dahal (Ph.D. ’20) and a team of UMD and FDA researchers to develop a novel technique to use small-angle X-ray scattering (SAXS) to estimate amyloid burden in the brain.
The team tapped SAXS for its ability to identify molecular structures based on their scattering patterns. Although SAXS is typically limited to the study of thin samples, the research group developed a way to apply a polychromatic X-ray beam and a 2-D spectroscopic detector to overcome the sample thickness limit of SAXS.
Early testing shows that their technique—known as spectral SAXS—can be used to effectively and noninvasively detect embedded targets in up to 5-cm-thick objects and in mouse models, all without the use of contrast agents. Their long-term hope is that one day clinicians will use this technique to assess how a patient’s Alzheimer’s disease has progressed by conducting an in-office scan of the brain that would take only a few minutes.
“With this spectral SAXS technology, we are aiming for histology-grade accuracy in terms of estimating amyloid burden in live animals, all within a matter of minutes and without using a contrast agent,” said Eshan Dahal, first author of the Scientific Reports paper. “We demonstrated the method by studying a well-established Alzheimer’s disease mouse model with amyloid pathology in just five minutes per studied location in the brain without any special sample preparation needs or injection of contrast agents. With further research and development, there may be an opportunity to expand this method in both preclinical and clinical settings to estimate amyloid burden or any protein aggregates of interest.”
Source: Read Full Article
