The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) envelope (E) protein has a C-terminus with a PDZ binding motif (PBM) that targets PDZ (PSD-95/Dlg/ZO-1) domains. This motif is identical to the SARS-CoV PBM.
In a recent study published on the bioRxiv* preprint server, ten human PDZ-containing proteins were identified, which showed significant interactions with the SARS-CoV-2 E protein in a PBM-dependent manner.
Study: Interactions of SARS-CoV-2 protein E with cell junctions and polarity PDZ-containing proteins. Image Credit: Orpheus FX / Shutterstock.com
About the study
In the current study, the researchers investigated the in vitro interactions of five PDZ domains including ZO-1, LNX2, PARD3, MLLT4, and MPP5 with the full-length E protein in a PBM-dependent manner. They also studied in cellulo the role of the full-length E protein in colocalization and sequestration of PDZ domains to the Golgi compartment.
The researchers further characterized the three crystal complex structures formed by PDZ/SARS-CoV-2 E with the LNX2, MLLT4, and MPP5 proteins and focused on their specific binding modes. They also compared the binding affinities of the SARS-CoV-2 Beta variant E protein's C-terminal and SARS-CoV-2 wild-type C-terminal with the PDZ domains.
Study findings
To determine the interaction between the selected five host PDZ domains and full-length E protein of SARS-CoV-2, the researchers tagged the five PDZ domains with glutathione S-transferase (GST) and tagged the full-length E protein with green fluorescence protein (GFP). The results indicated a PBM-dependent interaction between the PDZ domains of ZO-1, LNX2, MLLT4, and MPP5 and the full-length E protein of the wild-type virus with varying binding intensities, with PARD3 being the only exception.
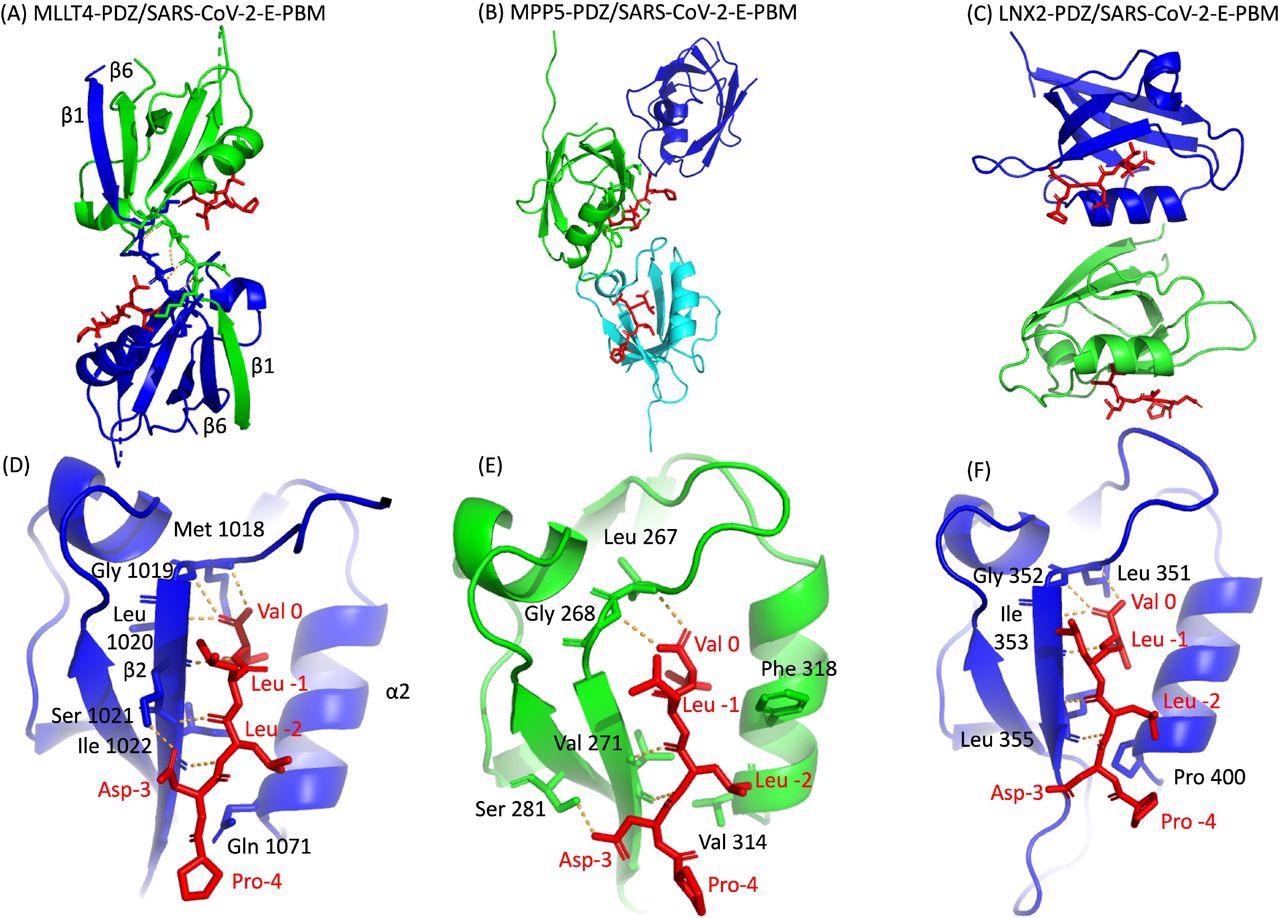
The researchers used a molecular replacement technique to characterize the crystal complex structure formed by the C-terminal sequence of the E protein by PBM-dependent interaction with the PDZ domains of MPP5, MLLT4, and LNX2. The PDZ domains of MPP5, MLLT4, and LNX2 bound to SARS-CoV-2-E PBM peptides with similar binding modes at positions 0 and -2.
Certainly, the N-terminal of the α2 helix is occupied by valine (Val 314), glutamine (Gln 1071), and proline (Pro 400) in PDZ domains of MPP5, MLLT4, and LNX2, respectively. The position -3 of the aspartic acid side chain forms hydrogen (H-bond) with PDZ domains of MPP5 and MLLT4; however, no interaction of proline at position -4 was found with any of the PDZ domains.
In the in cellulo study, the transfection of the SARS-CoV-2 E protein with GFP alone showed no accumulation of GFP-tagged PDZ domains. However, when GFP was co-transfected with ALFA-E, the SARS-CoV-2 E protein was involved in the relocalization and recruitment of PDZ domains from ZO1, MLLT4, MPP5, LNX2, and PARD3 into the Golgi compartment. This indicates that the SARS-CoV-2-E protein can potentially bind to PDZ domains in a PBM-dependent manner.
Binding affinities of PDZ domains
The current study determined the binding affinity of the original SARS-CoV-2 E protein C-terminal PBM peptide and the Beta variant P71L PBM peptide for the PDZ domains of ZO1, MLLT4, MPP5, LNX2, and PARD3 using the microscale thermophoresis technique.
For the original SARS-CoV-2 E peptide, the PDZ of MPP5 and the PDZ2 of ZO-1 displayed the best affinities with Kd values of 30 micromolar (μM) and 15 μM, respectively. The MLLT4-PDZ and PARD3-PDZ3 affinities were lower, with Kd values of 569 µM and 341 µM, respectively.
For the Beta variant P71L mutation, the affinities for the PDZ domains of MLLT4 and PARD3 were not detectable in the tested concentration range, thereby suggesting that they might not react with PDZ domains of MLLT4 and PARD3 or else the binding affinities were beyond the tested concentration range. The mutation P71L had no significant impact on the binding affinity of MPP5 but significantly reduced the affinity for ZO-1-PDZ2 with a Kd of 133 µM. The affinity of LNX2-PDZ2 improved from 289 µM to 47 µM in mutant P71L.
Overall, the results show that the four PDZ domains MPP5, ZO-1, MLLT4, and PARD3 exhibit different binding affinities for P71L. This suggests a differential specificity profile of the original C-terminal SARS-CoV-2 E protein PBM and the Beta C-terminal SARS-CoV-2 E protein PBM P71L for the PDZ domain.
*Important notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Zhu, Y., Alvarez, F., Wolff, N., et al. (2021). Interactions of SARS-CoV-2 protein E with cell junctions and polarity PDZ-containing proteins. bioRxiv. doi:10.1101/2021.12.04.471219. https://www.biorxiv.org/content/10.1101/2021.12.04.471219v1.
Posted in: Molecular & Structural Biology | Medical Science News | Medical Research News | Disease/Infection News
Tags: Aspartic Acid, binding affinity, Cell, Coronavirus, Coronavirus Disease COVID-19, Fluorescence, Glutamine, Helix, in vitro, Mutation, Peptides, Proline, Protein, Protein C, Respiratory, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Syndrome, Transfection, Valine, Virus

Written by
Susha Cheriyedath
Susha has a Bachelor of Science (B.Sc.) degree in Chemistry and Master of Science (M.Sc) degree in Biochemistry from the University of Calicut, India. She always had a keen interest in medical and health science. As part of her masters degree, she specialized in Biochemistry, with an emphasis on Microbiology, Physiology, Biotechnology, and Nutrition. In her spare time, she loves to cook up a storm in the kitchen with her super-messy baking experiments.
Source: Read Full Article
