New research led by Hana El-Samad of the University of California, San Francisco, and the Chan-Zuckerberg Biohub confirmed the beneficial use of an antiviral therapeutic called SARSNotch, which uses a combination of the SynNotch platform and de-novo designed protein binders. SARSNotch uses sentinel T cells to detect and mount a genetic response toward the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike protein.
The SynNotch platform had shown prior success with detecting the hepatitis B virus, and the research team wanted to expand on its use for the coronavirus. Moving forward, sentinel cells could be further expanded on for future drug development and therapeutic screenings.
"We demonstrate that these sentinel cells can detect purified Spike protein and a model of infected cells that are expressing Spike, and we show that this sensing approach is portable between different cell types, including primary human T lymphocytes and adherent cells. We suggest that this system, when coupled with appropriate therapeutic payloads, represents a potential next-generation targeted therapeutic against future pathogenic threats."
The study "Sentinel cells enable genetic detection of SARS-CoV-2 Spike protein" is available as a preprint on the bioRxiv* server, while the article undergoes peer review.
How they did it
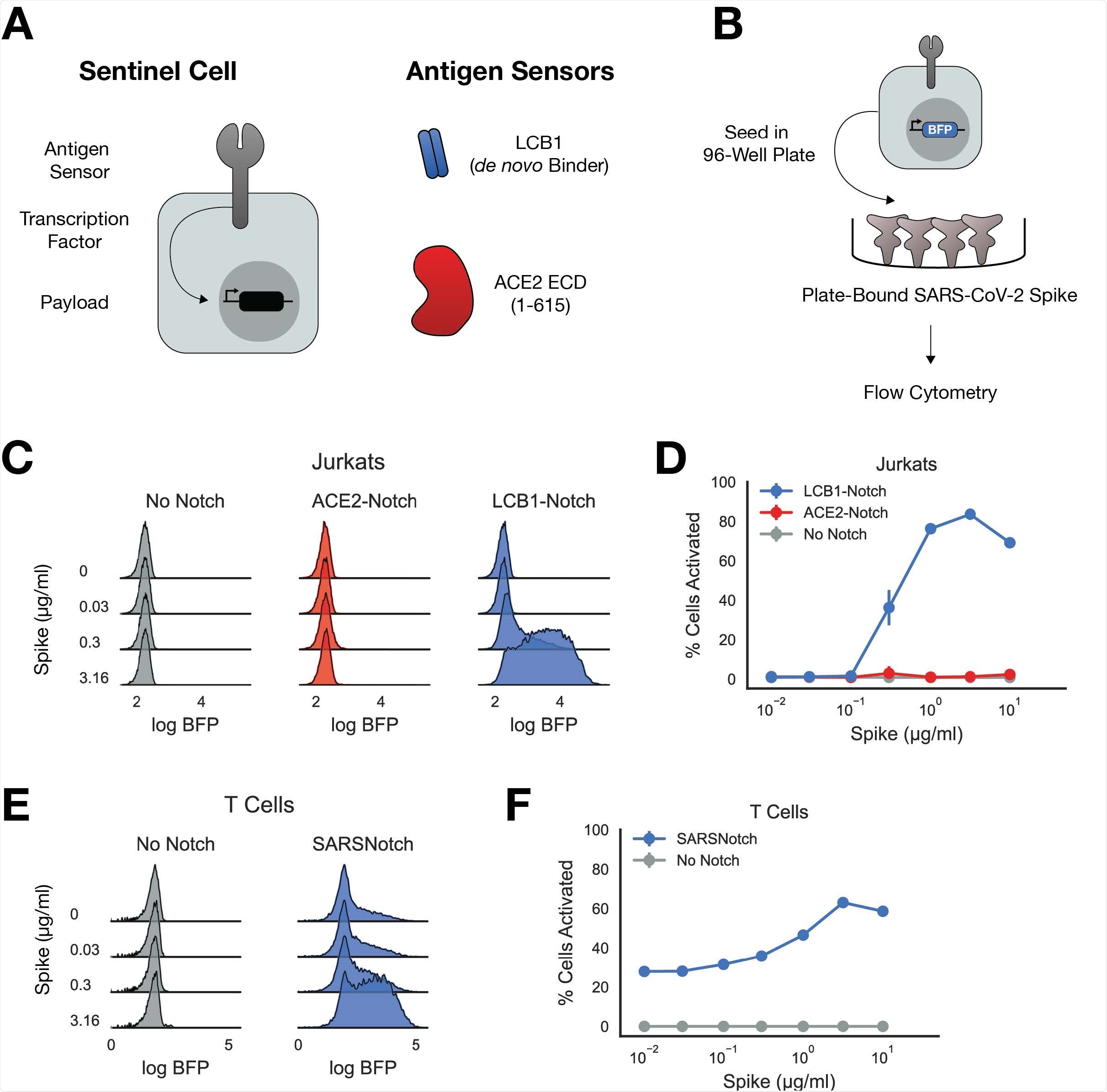
The researchers chose the SynNotch model because its platform allows for swappable antigen-sensing domains — making it adaptable for use viruses beyond SARS-CoV-2.
It is composed of an extracellular sensor domain that would target and recognize a specific epitope, fused to a user-defined cleavable intracellular domain retaining a transcription factor at the cellular membrane. Once the antigen is identified, the transcription factor is cleaved and moves into the nucleus to allow for output expression.
For SARS-CoV-2, they used the (5X)UAS-ybTATA as the transcriptional output and the ACE2 as the extracellular sensor. They also used de novo binders to serve as antigen sensors for the spike protein. Specifically, they screened for short protein sequences bound to the spike protein and from the screening used LCB1 as the antigen sensor.
SARSNotch showed high sensitivity in detecting spike protein
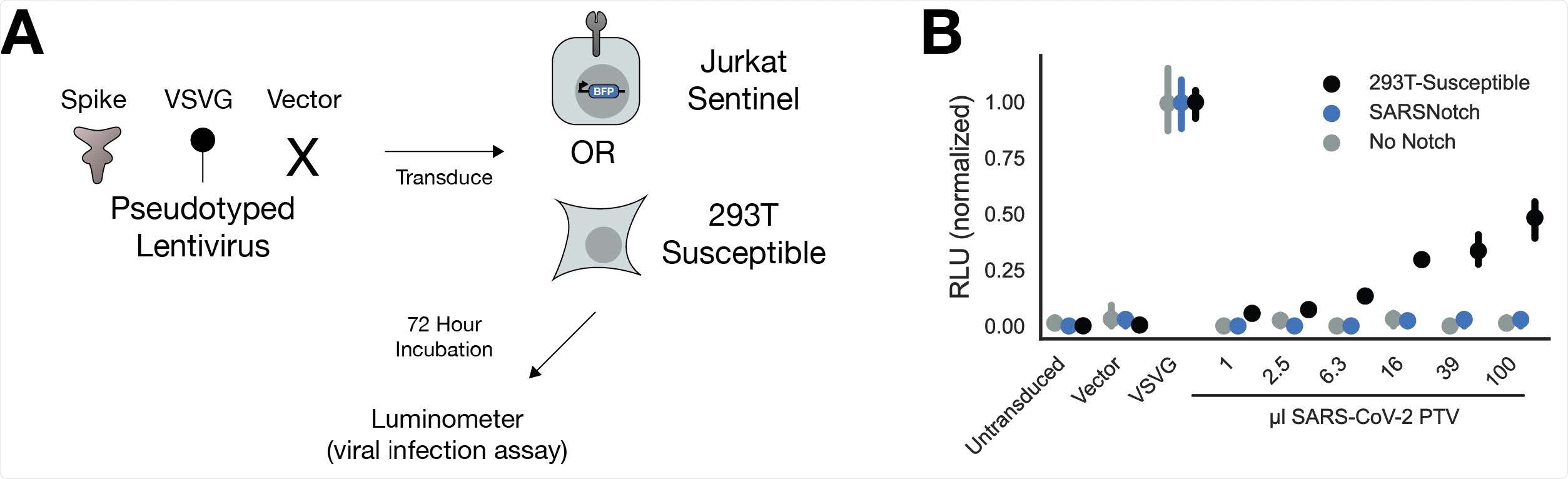
When testing the SARSNotch on pseudoviruses, they found sentinel cells successful in detecting the spike protein alone or when bound to another cell within 24 hours. Detection sensitivity was high with the platform detecting a purified spike protein at 0.3µg/ml.
They also confirmed the platform's versatility with it being able to be used between various cell types, including primary human T lymphocytes and adherent cells. Thus, the platform's application could be used for delivering therapeutics in a live cell-based therapy and in vitro.
There were some unexpected difficulties with using ACE2 as the antigen sensor as it did not provide a detectable signal output in the presence of the spike protein. The researchers suggest this may be due to size limitations in antigen sensors.
"Perhaps more likely, the 600 amino acid-long ACE2 extracellular domain is between 2 and 3 times the size of the scFvs and nanobodies that have been used as antigen sensors previously, suggesting potential limits on the size of antigen sensors that SynNotch can accommodate."
Given the adequate size of the de novo-designed binder, LCB1, being only 55 amino acids long, there is also the possibility of specific binding geometry needed for SynNotch activation. Future development on SynNotch would require experimenting with novel binders with a wide range of proteins as antigen sensors.
SARSNotch was able to detect the spike protein without increasing the risk of viral infection in sentinel cells. Sentinel cells appeared to not be a target for viral infection, adding strength to the platform.
Future of SARSNotch
There is still much work to be done before sentinel cells can be implemented for cell-based therapeutics in humans. Refining antigen sensors, reducing costs, and increasing the production rate of platforms are a few things that need to be modified in the future.
However, the researchers say that open collaboration from their scientific colleagues in refining the platform could serve as a promising therapy for detecting and neutralizing future viral threats.
*Important Notice
bioRxiv publishes preliminary scientific reports that are not peer-reviewed and, therefore, should not be regarded as conclusive, guide clinical practice/health-related behavior, or treated as established information.
- Weinberg Z et al. Sentinel cells enable genetic detection of SARS-CoV-2 Spike protein. bioRxiv, 2021. doi: https://doi.org/10.1101/2021.04.20.440678, https://www.biorxiv.org/content/10.1101/2021.04.20.440678v1
Posted in: Device / Technology News | Medical Research News | Disease/Infection News
Tags: ACE2, Amino Acid, Antigen, Cell, Cell Line, Coronavirus, Coronavirus Disease COVID-19, Cytometry, Flow Cytometry, Fluorescence, Fluorophore, Genetic, Hepatitis B, Hepatitis B Virus, in vitro, Intracellular, Luciferase, Nanobodies, Protein, Pseudovirus, Research, Respiratory, SARS, SARS-CoV-2, Severe Acute Respiratory, Severe Acute Respiratory Syndrome, Spike Protein, Syndrome, Therapeutics, Transcription, Virus

Written by
Jocelyn Solis-Moreira
Jocelyn Solis-Moreira graduated with a Bachelor's in Integrative Neuroscience, where she then pursued graduate research looking at the long-term effects of adolescent binge drinking on the brain's neurochemistry in adulthood.
Source: Read Full Article
